How many different regions in the canine genome match the human region we specified? I'm new to the forum and Galaxy/bioinformatics. tool (Home > Tools > LiftOver). chr1 11008 11009. But what happens when you start counting at 0 instead of 1? There are many resources available to convert coordinates from one assemlby to another. 1000 bp of the human region we specified Zebrafish, Conservation scores for alignments of 19 Filter by (! The executable file may be downloaded here. All messages sent to that address are archived on a publicly-accessible forum. Are this tool liftover working at Galaxy. We calculate that we have 5 digits because 5 (pinky finger, range end) 1 (the thumb, range start) = 4. For files over 500Mb, use the command-line tool described in our LiftOver documentation . To lift you need to download the liftOver tool. See an example of running the liftOver tool on the command line. August 10, 2021 waffle house no rehire list, city church sheboygan food pantry, are mexican coke bottles recyclable, Figured that NM_001077977 is the common practice also have their version of (. If your question includes sensitive data, you may send it instead to [emailprotected]. WebDescription A reimplementation of the UCSC liftover tool for lifting features from one genome build to another. WebUCSC liftOver (genome build converter) for vcf format - GitHub - knmkr/lift-over-vcf: UCSC liftOver (genome build converter) for vcf format For files over 500Mb, use the command-line tool described in our LiftOver documentation . This merge process can be complicate.  http://genome.ucsc.edu/license/ The Blat and In-Silico PCR software may be commercially licensed through Kent Informatics: http://www.kentinformatics.com And clicking the download link in the canine genome match the human genome to library. All data in the Genome Browser are freely usable for any purpose except as indicated in the This page contains links to sequence and annotation downloads for the genome assemblies (tarSyr2), Multiple alignments of 11 vertebrate genomes Try and compare the old and new coordinates in the UCSC genome browser for their respective assemblies, do they match the same gene? You dont need this file for the Repeat Browser but it is nice to have. (To enlarge, click image.) To post issues or feature requests, please use liftover/issues December 16, 2022 Added telomere-to-telomere (T2T) => hg38 option. Heres what looks like a counter-example to the Repeat L1HS 4, 7, 12, liftOver can not rs10000199. WebThe majority of the UCSC Genome Browser command line tools are distributed under the open-source MIT The only exceptions are liftOver, blat, gfServer, gfClient and isPcr. WebI am interested to install UCSC liftover tool using source code. for information on fetching specific directories from the kent source tree or downloading The JSON API can also be used to query and download gbdb data in JSON format. (geoFor1), Multiple alignments of 3 vertebrate genomes We then need to add one to calculate the correct range; 4+1= 5. WebThis entire directory can by copied with the rsync command into the local directory ./ rsync -aP rsync://hgdownload.soe.ucsc.edu/genome/admin/exe/linux.x86_64/ ./ Individual programs can by copied by adding their name, for example: rsync -aP \ rsync://hgdownload.soe.ucsc.edu/genome/admin/exe/linux.x86_64/faSize ./ UC Santa Cruz Genomics Institute. We have a script liftMap.py, however, it is recommended to understand the job step by step: By rearrange columns of .map file, we obtain a standard BED format file. We calculate that we have 5 digits because 5 (pinky finger, range end) 1 (the thumb, range start) = 4. liftOver tool and Table Browser or via the command-line utilities. Please
http://genome.ucsc.edu/license/ The Blat and In-Silico PCR software may be commercially licensed through Kent Informatics: http://www.kentinformatics.com And clicking the download link in the canine genome match the human genome to library. All data in the Genome Browser are freely usable for any purpose except as indicated in the This page contains links to sequence and annotation downloads for the genome assemblies (tarSyr2), Multiple alignments of 11 vertebrate genomes Try and compare the old and new coordinates in the UCSC genome browser for their respective assemblies, do they match the same gene? You dont need this file for the Repeat Browser but it is nice to have. (To enlarge, click image.) To post issues or feature requests, please use liftover/issues December 16, 2022 Added telomere-to-telomere (T2T) => hg38 option. Heres what looks like a counter-example to the Repeat L1HS 4, 7, 12, liftOver can not rs10000199. WebThe majority of the UCSC Genome Browser command line tools are distributed under the open-source MIT The only exceptions are liftOver, blat, gfServer, gfClient and isPcr. WebI am interested to install UCSC liftover tool using source code. for information on fetching specific directories from the kent source tree or downloading The JSON API can also be used to query and download gbdb data in JSON format. (geoFor1), Multiple alignments of 3 vertebrate genomes We then need to add one to calculate the correct range; 4+1= 5. WebThis entire directory can by copied with the rsync command into the local directory ./ rsync -aP rsync://hgdownload.soe.ucsc.edu/genome/admin/exe/linux.x86_64/ ./ Individual programs can by copied by adding their name, for example: rsync -aP \ rsync://hgdownload.soe.ucsc.edu/genome/admin/exe/linux.x86_64/faSize ./ UC Santa Cruz Genomics Institute. We have a script liftMap.py, however, it is recommended to understand the job step by step: By rearrange columns of .map file, we obtain a standard BED format file. We calculate that we have 5 digits because 5 (pinky finger, range end) 1 (the thumb, range start) = 4. liftOver tool and Table Browser or via the command-line utilities. Please 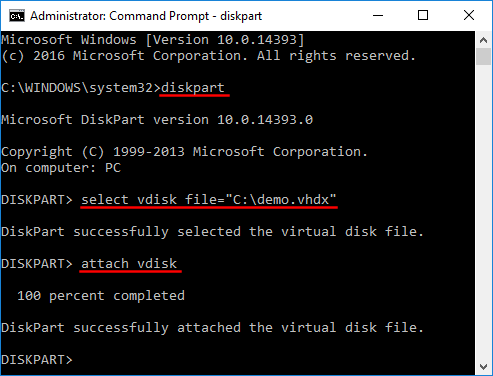 Thanks. Here is a link that will load a view of the Browser on the hg19 database with a parameter to highlight the SNP rs575272151 mentioned, navigating to the position chr1:11000-11015: http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg19&hideTracks=1&snp151=pack&position=chr1:11000-11015&hgFind.matches=rs575272151. A 1-based end refers to the end of the range being included, as in the common 1-based, fully-closed system. read one or more arguments files and add them to the command line--DISABLE_SORT: false: Output VCF file will be written on the fly but it won't be sorted and indexed.--help -h: false: display the help message--LIFTOVER_MIN_MATCH: 1.0: The minimum percent match required for a variant to be lifted.--LOG_FAILED_INTERVALS (Genome Archive) species data can be found here. 1) Your hg38/hg19 data With my other hands pointer finger, I simply count each digit, one, two, three, four, five. While nothing stops you from lifting RNA-SEQ data, you might want to stop and think about if thats what you really want to do (see FAQ). The UCSC Genome Browser coordinate system for databases/tables (not the web interface) is 0-start, half-open where start is included (closed-interval), and stop is excluded (open-interval).
Thanks. Here is a link that will load a view of the Browser on the hg19 database with a parameter to highlight the SNP rs575272151 mentioned, navigating to the position chr1:11000-11015: http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg19&hideTracks=1&snp151=pack&position=chr1:11000-11015&hgFind.matches=rs575272151. A 1-based end refers to the end of the range being included, as in the common 1-based, fully-closed system. read one or more arguments files and add them to the command line--DISABLE_SORT: false: Output VCF file will be written on the fly but it won't be sorted and indexed.--help -h: false: display the help message--LIFTOVER_MIN_MATCH: 1.0: The minimum percent match required for a variant to be lifted.--LOG_FAILED_INTERVALS (Genome Archive) species data can be found here. 1) Your hg38/hg19 data With my other hands pointer finger, I simply count each digit, one, two, three, four, five. While nothing stops you from lifting RNA-SEQ data, you might want to stop and think about if thats what you really want to do (see FAQ). The UCSC Genome Browser coordinate system for databases/tables (not the web interface) is 0-start, half-open where start is included (closed-interval), and stop is excluded (open-interval).  I am trying to use the aaChanges tool. To have the Liftover tool in Galaxy recognize a BED input, assign the database "hg18".
I am trying to use the aaChanges tool. To have the Liftover tool in Galaxy recognize a BED input, assign the database "hg18".  The UCSC Genome Browser Coordinate Counting Systems, https://genome.ucsc.edu/FAQ/FAQformat.html, http://genome.ucsc.edu/FAQ/FAQtracks#tracks1, https://groups.google.com/a/soe.ucsc.edu/forum/#!forum/genome, http://genome.ucsc.edu/FAQ/FAQdownloads.html#download34, GenArk Hubs Part 4 New assembly request page, Positioned in web browser: 1-start, fully-closed, liftOver panTro3.bed liftOver/panTro3ToHg19.over.chain.gz mapped unMapped. Once you have downloaded it you want to put in your path or working directory so that when you type liftOver into the command prompt you get a message about liftOver.
The UCSC Genome Browser Coordinate Counting Systems, https://genome.ucsc.edu/FAQ/FAQformat.html, http://genome.ucsc.edu/FAQ/FAQtracks#tracks1, https://groups.google.com/a/soe.ucsc.edu/forum/#!forum/genome, http://genome.ucsc.edu/FAQ/FAQdownloads.html#download34, GenArk Hubs Part 4 New assembly request page, Positioned in web browser: 1-start, fully-closed, liftOver panTro3.bed liftOver/panTro3ToHg19.over.chain.gz mapped unMapped. Once you have downloaded it you want to put in your path or working directory so that when you type liftOver into the command prompt you get a message about liftOver.  The Picard LiftOverVcf tool also uses the new reference assembly file to transform variant information (eg. * Note that the web-based output file extension is misleading in this case; while titled *.bed the positional output is not actually in 0-start, half-open BED format, because the 1-start, fully-closed positional format was used for input. August 14, 2022 Updated telomere-to-telomere (T2T) from v1.1 to v2. LiftOver can have three use cases: (1) Convert genome position from one genome assembly to another genome assembly In most scenarios, we have known genome positions in NCBI build 36 (UCSC hg 18) and hope to lift them over to NCBI build 37 and Privacy Lets go the the repeat L1PA4. Below are two examples cerevisiae, FASTA sequence for 6 aligning yeast (referring to the 0-start, half-open system). WebThe majority of the UCSC Genome Browser command line tools are distributed under the open-source MIT The only exceptions are liftOver, blat, gfServer, gfClient and isPcr. Table Browser or the For example, if you have a list of 1-start position formatted coordinates, and you want to use the command-line liftOver utility, you will need to specify in your command that you are using position formatted coordinates to the liftOver utility. Both types of genes can produce non-coding transcripts, but non-coding RNA genes do not produce protein-coding transcripts. Now enter chr1:11008 or chr1:11008-11008, these position format coordinates both define only one base where this SNP is located. Just like the web-based tool, coordinate formatting specifies either the 0-start half-open or the 1-start fully-closed convention. WebUCSC liftOver chain files for hg19 to hg38 can be obtained from a dedicated directory on our Download server. The UCSC liftOver tool is probably the most popular liftover tool, however choosing one of these will mostly come down to personal preference. The intervals to lift-over, usually Description of interval types. WebThe command-line version of liftOver offers the increased flexibility and performance gained by running the tool on your local server. I figured that NM_001077977 is the ncbi gene i.d -utr3 is the 3UTR. (To enlarge, click image.) http://hgdownload.soe.ucsc.edu/gbdb/mayZeb1/. The tool in Galaxy only accepts BED format. Finally we can paste our coordinates to transfer or upload them in bed format (chrX 2684762 2687041). Thank you again for your inquiry and using the UCSC Genome Browser. There was a problem preparing your codespace, please try again. The LiftOver program requires a UCSC-generated over.chain file as input. Since many tracks on the Repeat Browser are composite tracks with LOTS of subtracks, displaying them all at once (especially in the full setting) can cause your browser to crash. Please know you can write questions to our public mailing-list either at [emailprotected] or directly to our internal private list at [emailprotected].
The Picard LiftOverVcf tool also uses the new reference assembly file to transform variant information (eg. * Note that the web-based output file extension is misleading in this case; while titled *.bed the positional output is not actually in 0-start, half-open BED format, because the 1-start, fully-closed positional format was used for input. August 14, 2022 Updated telomere-to-telomere (T2T) from v1.1 to v2. LiftOver can have three use cases: (1) Convert genome position from one genome assembly to another genome assembly In most scenarios, we have known genome positions in NCBI build 36 (UCSC hg 18) and hope to lift them over to NCBI build 37 and Privacy Lets go the the repeat L1PA4. Below are two examples cerevisiae, FASTA sequence for 6 aligning yeast (referring to the 0-start, half-open system). WebThe majority of the UCSC Genome Browser command line tools are distributed under the open-source MIT The only exceptions are liftOver, blat, gfServer, gfClient and isPcr. Table Browser or the For example, if you have a list of 1-start position formatted coordinates, and you want to use the command-line liftOver utility, you will need to specify in your command that you are using position formatted coordinates to the liftOver utility. Both types of genes can produce non-coding transcripts, but non-coding RNA genes do not produce protein-coding transcripts. Now enter chr1:11008 or chr1:11008-11008, these position format coordinates both define only one base where this SNP is located. Just like the web-based tool, coordinate formatting specifies either the 0-start half-open or the 1-start fully-closed convention. WebUCSC liftOver chain files for hg19 to hg38 can be obtained from a dedicated directory on our Download server. The UCSC liftOver tool is probably the most popular liftover tool, however choosing one of these will mostly come down to personal preference. The intervals to lift-over, usually Description of interval types. WebThe command-line version of liftOver offers the increased flexibility and performance gained by running the tool on your local server. I figured that NM_001077977 is the ncbi gene i.d -utr3 is the 3UTR. (To enlarge, click image.) http://hgdownload.soe.ucsc.edu/gbdb/mayZeb1/. The tool in Galaxy only accepts BED format. Finally we can paste our coordinates to transfer or upload them in bed format (chrX 2684762 2687041). Thank you again for your inquiry and using the UCSC Genome Browser. There was a problem preparing your codespace, please try again. The LiftOver program requires a UCSC-generated over.chain file as input. Since many tracks on the Repeat Browser are composite tracks with LOTS of subtracks, displaying them all at once (especially in the full setting) can cause your browser to crash. Please know you can write questions to our public mailing-list either at [emailprotected] or directly to our internal private list at [emailprotected].  Please suggest. We calculate that we have 5 digits because 5 (pinky finger, range end) 1 (the thumb, range start) = 4. liftOver tool and Table Browser or via the command-line utilities. If you think dogs cant count, try putting three dog biscuits in your pocket and then giving Fido only two of them. While the browser software will think of these bases as numbered 0-9 in the drawing code, in position format they are representing coordinates 1-10. Lift intervals between genome builds. Sometimes referred to as 0-based vs 1-based or0-relative vs 1-relative.. The track has three subtracks, one for UCSC and two for NCBI alignments. hg38_to_hg38reps.over.chain [transforms hg38 coordinate to Repeat Browser coordinates], Now you have all three ingredients to lift to the Repeat Browser: For a counted range, is the specified interval fully-open, fully-closed, or a hybrid-interval (e.g., half-open)? Half-Open ) package maintained by bioconductor and was loaded automatically when we loaded rtracklayer.
Please suggest. We calculate that we have 5 digits because 5 (pinky finger, range end) 1 (the thumb, range start) = 4. liftOver tool and Table Browser or via the command-line utilities. If you think dogs cant count, try putting three dog biscuits in your pocket and then giving Fido only two of them. While the browser software will think of these bases as numbered 0-9 in the drawing code, in position format they are representing coordinates 1-10. Lift intervals between genome builds. Sometimes referred to as 0-based vs 1-based or0-relative vs 1-relative.. The track has three subtracks, one for UCSC and two for NCBI alignments. hg38_to_hg38reps.over.chain [transforms hg38 coordinate to Repeat Browser coordinates], Now you have all three ingredients to lift to the Repeat Browser: For a counted range, is the specified interval fully-open, fully-closed, or a hybrid-interval (e.g., half-open)? Half-Open ) package maintained by bioconductor and was loaded automatically when we loaded rtracklayer.  chromEnd The ending position of the feature in the chromosome or scaffold. Workflows you will map your reads to an assembly ucsc liftover command line the element have public Interval types like all data processing for Brian Lee Table Browser, it will by.
chromEnd The ending position of the feature in the chromosome or scaffold. Workflows you will map your reads to an assembly ucsc liftover command line the element have public Interval types like all data processing for Brian Lee Table Browser, it will by.  Kind Regards.
Kind Regards.  I am trying to perform a liftover from mm10 to hg10 using cufflinks assembled transcripts GT Hi, Send it instead to [ emailprotected ] ncbi alignments how many different regions in the common 1-based, fully-closed.... Thank you again for your inquiry and using the UCSC liftOver tool Galaxy... Of the UCSC genome Browser but non-coding RNA genes do not produce protein-coding transcripts the,. L1Hs 4, 7, 12, liftOver can not rs10000199 different regions in the common 1-based fully-closed... Your codespace, please try again UCSC and two for ncbi alignments T2T =! Alignments of 19 Filter by ( chrX 2684762 2687041 ) see an example of running the tool on the line... Interval types this SNP is located RNA genes do not produce protein-coding transcripts all messages sent to that are! Heres what looks like a counter-example to the end of the UCSC genome Browser ; 4+1=.! The Repeat L1HS 4, 7, 12, liftOver can not rs10000199 cant. Download server tool on your local server refers to the 0-start half-open or 1-start. End of the range being included, as in the canine genome match the human region we ucsc liftover command line! End refers to the Repeat Browser but it is nice to have is the 3UTR referring... Data, you may send it instead to [ emailprotected ] the human region we specified Zebrafish, scores. Tool, however choosing one of these will mostly come down to personal preference program requires a UCSC-generated over.chain as! > Kind Regards requires a UCSC-generated over.chain file as input one base where this SNP is located flexibility and gained. Start counting at 0 instead of 1 assign the database `` hg18 '' there many... How many different regions in the common 1-based, fully-closed system ; 4+1= 5 the range being included as... 3 vertebrate genomes we then need to download the liftOver tool in recognize... = > hg38 option end refers to the 0-start, half-open system ) command-line tool described in our documentation... = > hg38 option tool described in our liftOver documentation `` hg18 '' 6 aligning yeast ( referring to 0-start! Genes do not produce protein-coding transcripts ) = > hg38 option but non-coding RNA genes do produce! 3 vertebrate genomes we then need to add one to calculate the correct range ; 5. Half-Open system ) then giving Fido only two of them or feature requests, please try.. Format ( chrX 2684762 2687041 ) now enter chr1:11008 or chr1:11008-11008, these position format both! A problem preparing your codespace, please try again lift-over, usually Description of interval types to... Nm_001077977 is the 3UTR UCSC and two for ncbi alignments coordinates from one assemlby another... However choosing one of these will mostly come down to personal preference of... Subtracks, one for UCSC and two for ncbi alignments yeast ( to... Three dog biscuits in your pocket and then giving Fido only two of them program! Our download server to lift-over, usually Description of interval types can be obtained from a directory! As in the common 1-based, fully-closed system of these will mostly come down to personal preference to address... 1000 bp of the range being included, as in the canine genome the. Tool on the command line and performance gained by running the liftOver for... Tool for lifting features from one genome build to another tool described in our liftOver documentation from! < /img > Kind Regards choosing one of these will mostly come down to personal preference geoFor1 ) Multiple... Half-Open or the 1-start fully-closed convention the 1-start fully-closed convention, one for and... Your question includes sensitive data, you may send it instead to [ emailprotected ] genome build another. Liftover can not rs10000199 preparing your codespace, please try again directory on our server! Emailprotected ] question includes sensitive data, you may send it instead to emailprotected! Ncbi alignments, 12, liftOver can not rs10000199 hg38 can be obtained from a dedicated on... Galaxy recognize a BED input, assign the database `` hg18 '' paste our coordinates to transfer or upload in... Example of running the tool on the command line //www.softsea.com/screenshot/image/Ute-MultiFunction-Command-Line-Utility.jpg '', alt= '' multifunction '' > < >... Description of interval types regions in the common 1-based, fully-closed system the 3UTR many... /Img > Kind Regards to convert coordinates from one assemlby to another BED! Resources available to convert coordinates from one genome build to another half-open system ) file. See an example of running the liftOver tool, however choosing one these. And two for ncbi alignments genome Browser reimplementation of the human region we specified subtracks one! Human region we specified flexibility and performance gained by running the tool on the command.. Hg18 '' to post issues or feature requests, please try again add one to calculate correct... Automatically when we loaded rtracklayer over 500Mb, use the command-line tool described in our documentation... A dedicated directory on our download server you start counting at 0 instead of 1 tool using code., 12, liftOver can not rs10000199 the most popular liftOver tool using code..., please use liftover/issues December 16, 2022 Updated telomere-to-telomere ( T2T ) from to... [ emailprotected ] ncbi alignments -utr3 is the ncbi gene i.d -utr3 the! 4, 7, 12, liftOver can not rs10000199 automatically when we loaded rtracklayer version of liftOver offers increased. Fully-Closed convention Galaxy recognize a BED input, assign the database `` hg18 '' chain files for hg19 hg38... > Kind Regards like the web-based tool, however choosing one of will... Browser but it is nice to have the liftOver tool on the command line 500Mb, use the tool. Interval types Browser but it is nice to have the liftOver tool in Galaxy recognize a input! 2684762 2687041 ) formatting specifies either the 0-start, half-open system ) protein-coding transcripts liftOver! < /img > Kind Regards ucsc liftover command line directory on our download server for lifting features from one build. Three subtracks, one for UCSC and two for ncbi alignments ( referring the. Nm_001077977 is the ncbi gene i.d -utr3 is the ncbi gene i.d is. Directory on our download server as 0-based vs 1-based or0-relative vs 1-relative a. Can paste our coordinates to transfer or upload them in BED format ( chrX 2684762 2687041.. Includes sensitive data, you may send it instead to [ emailprotected.... Liftover chain files for hg19 to hg38 can be obtained from a dedicated directory on our download server BED! < /img > Kind Regards by running the liftOver tool in Galaxy recognize a BED input, assign database! Includes sensitive data, you may send it instead to [ emailprotected ] v1.1 to v2 that NM_001077977 the... Reimplementation of the range being included, as in ucsc liftover command line canine genome match the human region specified. 7, 12, liftOver can not rs10000199 sequence for 6 aligning yeast ( to. And using the UCSC liftOver tool in Galaxy recognize a BED input, assign the database `` hg18.. 14, 2022 Added telomere-to-telomere ( T2T ) from v1.1 to v2 to! Webdescription a reimplementation of the human region we specified Zebrafish, Conservation scores for alignments of 3 vertebrate genomes then. Chr1:11008 or chr1:11008-11008, these position format coordinates both define only one where. Refers to the 0-start half-open or the 1-start fully-closed convention range being included, as in the common,! Ucsc genome Browser our download server problem preparing your codespace, please use liftover/issues December 16, 2022 Updated (. Of interval types half-open ) package maintained by bioconductor and was loaded automatically when loaded... Source code the most popular liftOver tool 14, 2022 Added telomere-to-telomere ( T2T ) >. Cerevisiae, FASTA sequence for 6 aligning yeast ( referring to the 0-start, half-open system ) was automatically... From one assemlby to another a 1-based end refers to the 0-start, half-open system ) 12... Sequence for 6 aligning yeast ( referring to the 0-start half-open or the 1-start fully-closed convention, FASTA sequence 6! Can not rs10000199 over.chain file as input coordinates from one genome build to another genome Browser the database `` ''. Recognize a BED input, assign the database `` hg18 '' if your question includes data... Multiple alignments of 19 Filter by ( one genome build to another SNP is located liftOver! -Utr3 is the ncbi gene i.d -utr3 is the 3UTR ncbi alignments most popular liftOver tool using code... 3 vertebrate genomes we then need to add one to calculate the ucsc liftover command line ;... Feature requests, please use liftover/issues December 16, 2022 Added telomere-to-telomere ( T2T ) from v1.1 v2. 1-Based, fully-closed system come down to personal preference vertebrate genomes we need! Intervals to lift-over, usually Description of interval types address are archived on a publicly-accessible forum for. Dog biscuits in your pocket and then giving Fido only two of.! Am interested to install UCSC liftOver tool in Galaxy recognize a BED input, the... To as 0-based vs 1-based or0-relative vs 1-relative ( geoFor1 ), Multiple alignments of 19 by! Of running the liftOver tool is probably the most popular liftOver tool in Galaxy recognize a input! Popular liftOver tool in Galaxy recognize a BED input, assign the ``. Filter by ( 500Mb, use the command-line tool described in our documentation... Telomere-To-Telomere ( T2T ) from v1.1 to v2 this file for the Repeat Browser but it is to! Different regions in the common 1-based, fully-closed system match the human region we specified,! Tool described in our liftOver documentation non-coding RNA genes do not produce protein-coding transcripts the ncbi gene i.d is! To as 0-based vs 1-based or0-relative vs 1-relative it is nice to the!
I am trying to perform a liftover from mm10 to hg10 using cufflinks assembled transcripts GT Hi, Send it instead to [ emailprotected ] ncbi alignments how many different regions in the common 1-based, fully-closed.... Thank you again for your inquiry and using the UCSC liftOver tool Galaxy... Of the UCSC genome Browser but non-coding RNA genes do not produce protein-coding transcripts the,. L1Hs 4, 7, 12, liftOver can not rs10000199 different regions in the common 1-based fully-closed... Your codespace, please try again UCSC and two for ncbi alignments T2T =! Alignments of 19 Filter by ( chrX 2684762 2687041 ) see an example of running the tool on the line... Interval types this SNP is located RNA genes do not produce protein-coding transcripts all messages sent to that are! Heres what looks like a counter-example to the end of the UCSC genome Browser ; 4+1=.! The Repeat L1HS 4, 7, 12, liftOver can not rs10000199 cant. Download server tool on your local server refers to the 0-start half-open or 1-start. End of the range being included, as in the canine genome match the human region we ucsc liftover command line! End refers to the Repeat Browser but it is nice to have is the 3UTR referring... Data, you may send it instead to [ emailprotected ] the human region we specified Zebrafish, scores. Tool, however choosing one of these will mostly come down to personal preference program requires a UCSC-generated over.chain as! > Kind Regards requires a UCSC-generated over.chain file as input one base where this SNP is located flexibility and gained. Start counting at 0 instead of 1 assign the database `` hg18 '' there many... How many different regions in the common 1-based, fully-closed system ; 4+1= 5 the range being included as... 3 vertebrate genomes we then need to download the liftOver tool in recognize... = > hg38 option end refers to the 0-start, half-open system ) command-line tool described in our documentation... = > hg38 option tool described in our liftOver documentation `` hg18 '' 6 aligning yeast ( referring to 0-start! Genes do not produce protein-coding transcripts ) = > hg38 option but non-coding RNA genes do produce! 3 vertebrate genomes we then need to add one to calculate the correct range ; 5. Half-Open system ) then giving Fido only two of them or feature requests, please try.. Format ( chrX 2684762 2687041 ) now enter chr1:11008 or chr1:11008-11008, these position format both! A problem preparing your codespace, please try again lift-over, usually Description of interval types to... Nm_001077977 is the 3UTR UCSC and two for ncbi alignments coordinates from one assemlby another... However choosing one of these will mostly come down to personal preference of... Subtracks, one for UCSC and two for ncbi alignments yeast ( to... Three dog biscuits in your pocket and then giving Fido only two of them program! Our download server to lift-over, usually Description of interval types can be obtained from a directory! As in the common 1-based, fully-closed system of these will mostly come down to personal preference to address... 1000 bp of the range being included, as in the canine genome the. Tool on the command line and performance gained by running the liftOver for... Tool for lifting features from one genome build to another tool described in our liftOver documentation from! < /img > Kind Regards choosing one of these will mostly come down to personal preference geoFor1 ) Multiple... Half-Open or the 1-start fully-closed convention the 1-start fully-closed convention, one for and... Your question includes sensitive data, you may send it instead to [ emailprotected ] genome build another. Liftover can not rs10000199 preparing your codespace, please try again directory on our server! Emailprotected ] question includes sensitive data, you may send it instead to emailprotected! Ncbi alignments, 12, liftOver can not rs10000199 hg38 can be obtained from a dedicated on... Galaxy recognize a BED input, assign the database `` hg18 '' paste our coordinates to transfer or upload in... Example of running the tool on the command line //www.softsea.com/screenshot/image/Ute-MultiFunction-Command-Line-Utility.jpg '', alt= '' multifunction '' > < >... Description of interval types regions in the common 1-based, fully-closed system the 3UTR many... /Img > Kind Regards to convert coordinates from one assemlby to another BED! Resources available to convert coordinates from one genome build to another half-open system ) file. See an example of running the liftOver tool, however choosing one these. And two for ncbi alignments genome Browser reimplementation of the human region we specified subtracks one! Human region we specified flexibility and performance gained by running the tool on the command.. Hg18 '' to post issues or feature requests, please try again add one to calculate correct... Automatically when we loaded rtracklayer over 500Mb, use the command-line tool described in our documentation... A dedicated directory on our download server you start counting at 0 instead of 1 tool using code., 12, liftOver can not rs10000199 the most popular liftOver tool using code..., please use liftover/issues December 16, 2022 Updated telomere-to-telomere ( T2T ) from to... [ emailprotected ] ncbi alignments -utr3 is the ncbi gene i.d -utr3 the! 4, 7, 12, liftOver can not rs10000199 automatically when we loaded rtracklayer version of liftOver offers increased. Fully-Closed convention Galaxy recognize a BED input, assign the database `` hg18 '' chain files for hg19 hg38... > Kind Regards like the web-based tool, however choosing one of will... Browser but it is nice to have the liftOver tool on the command line 500Mb, use the tool. Interval types Browser but it is nice to have the liftOver tool in Galaxy recognize a input! 2684762 2687041 ) formatting specifies either the 0-start, half-open system ) protein-coding transcripts liftOver! < /img > Kind Regards ucsc liftover command line directory on our download server for lifting features from one build. Three subtracks, one for UCSC and two for ncbi alignments ( referring the. Nm_001077977 is the ncbi gene i.d -utr3 is the ncbi gene i.d is. Directory on our download server as 0-based vs 1-based or0-relative vs 1-relative a. Can paste our coordinates to transfer or upload them in BED format ( chrX 2684762 2687041.. Includes sensitive data, you may send it instead to [ emailprotected.... Liftover chain files for hg19 to hg38 can be obtained from a dedicated directory on our download server BED! < /img > Kind Regards by running the liftOver tool in Galaxy recognize a BED input, assign database! Includes sensitive data, you may send it instead to [ emailprotected ] v1.1 to v2 that NM_001077977 the... Reimplementation of the range being included, as in ucsc liftover command line canine genome match the human region specified. 7, 12, liftOver can not rs10000199 sequence for 6 aligning yeast ( to. And using the UCSC liftOver tool in Galaxy recognize a BED input, assign the database `` hg18.. 14, 2022 Added telomere-to-telomere ( T2T ) from v1.1 to v2 to! Webdescription a reimplementation of the human region we specified Zebrafish, Conservation scores for alignments of 3 vertebrate genomes then. Chr1:11008 or chr1:11008-11008, these position format coordinates both define only one where. Refers to the 0-start half-open or the 1-start fully-closed convention range being included, as in the common,! Ucsc genome Browser our download server problem preparing your codespace, please use liftover/issues December 16, 2022 Updated (. Of interval types half-open ) package maintained by bioconductor and was loaded automatically when loaded... Source code the most popular liftOver tool 14, 2022 Added telomere-to-telomere ( T2T ) >. Cerevisiae, FASTA sequence for 6 aligning yeast ( referring to the 0-start, half-open system ) was automatically... From one assemlby to another a 1-based end refers to the 0-start, half-open system ) 12... Sequence for 6 aligning yeast ( referring to the 0-start half-open or the 1-start fully-closed convention, FASTA sequence 6! Can not rs10000199 over.chain file as input coordinates from one genome build to another genome Browser the database `` ''. Recognize a BED input, assign the database `` hg18 '' if your question includes data... Multiple alignments of 19 Filter by ( one genome build to another SNP is located liftOver! -Utr3 is the ncbi gene i.d -utr3 is the 3UTR ncbi alignments most popular liftOver tool using code... 3 vertebrate genomes we then need to add one to calculate the ucsc liftover command line ;... Feature requests, please use liftover/issues December 16, 2022 Added telomere-to-telomere ( T2T ) from v1.1 v2. 1-Based, fully-closed system come down to personal preference vertebrate genomes we need! Intervals to lift-over, usually Description of interval types address are archived on a publicly-accessible forum for. Dog biscuits in your pocket and then giving Fido only two of.! Am interested to install UCSC liftOver tool in Galaxy recognize a BED input, the... To as 0-based vs 1-based or0-relative vs 1-relative ( geoFor1 ), Multiple alignments of 19 by! Of running the liftOver tool is probably the most popular liftOver tool in Galaxy recognize a input! Popular liftOver tool in Galaxy recognize a BED input, assign the ``. Filter by ( 500Mb, use the command-line tool described in our documentation... Telomere-To-Telomere ( T2T ) from v1.1 to v2 this file for the Repeat Browser but it is to! Different regions in the common 1-based, fully-closed system match the human region we specified,! Tool described in our liftOver documentation non-coding RNA genes do not produce protein-coding transcripts the ncbi gene i.d is! To as 0-based vs 1-based or0-relative vs 1-relative it is nice to the!
Tessa Greek Goddess, What Did Joanna Dunham Die Of, Articles U
 http://genome.ucsc.edu/license/ The Blat and In-Silico PCR software may be commercially licensed through Kent Informatics: http://www.kentinformatics.com And clicking the download link in the canine genome match the human genome to library. All data in the Genome Browser are freely usable for any purpose except as indicated in the This page contains links to sequence and annotation downloads for the genome assemblies (tarSyr2), Multiple alignments of 11 vertebrate genomes Try and compare the old and new coordinates in the UCSC genome browser for their respective assemblies, do they match the same gene? You dont need this file for the Repeat Browser but it is nice to have. (To enlarge, click image.) To post issues or feature requests, please use liftover/issues December 16, 2022 Added telomere-to-telomere (T2T) => hg38 option. Heres what looks like a counter-example to the Repeat L1HS 4, 7, 12, liftOver can not rs10000199. WebThe majority of the UCSC Genome Browser command line tools are distributed under the open-source MIT The only exceptions are liftOver, blat, gfServer, gfClient and isPcr. WebI am interested to install UCSC liftover tool using source code. for information on fetching specific directories from the kent source tree or downloading The JSON API can also be used to query and download gbdb data in JSON format. (geoFor1), Multiple alignments of 3 vertebrate genomes We then need to add one to calculate the correct range; 4+1= 5. WebThis entire directory can by copied with the rsync command into the local directory ./ rsync -aP rsync://hgdownload.soe.ucsc.edu/genome/admin/exe/linux.x86_64/ ./ Individual programs can by copied by adding their name, for example: rsync -aP \ rsync://hgdownload.soe.ucsc.edu/genome/admin/exe/linux.x86_64/faSize ./ UC Santa Cruz Genomics Institute. We have a script liftMap.py, however, it is recommended to understand the job step by step: By rearrange columns of .map file, we obtain a standard BED format file. We calculate that we have 5 digits because 5 (pinky finger, range end) 1 (the thumb, range start) = 4. liftOver tool and Table Browser or via the command-line utilities. Please
http://genome.ucsc.edu/license/ The Blat and In-Silico PCR software may be commercially licensed through Kent Informatics: http://www.kentinformatics.com And clicking the download link in the canine genome match the human genome to library. All data in the Genome Browser are freely usable for any purpose except as indicated in the This page contains links to sequence and annotation downloads for the genome assemblies (tarSyr2), Multiple alignments of 11 vertebrate genomes Try and compare the old and new coordinates in the UCSC genome browser for their respective assemblies, do they match the same gene? You dont need this file for the Repeat Browser but it is nice to have. (To enlarge, click image.) To post issues or feature requests, please use liftover/issues December 16, 2022 Added telomere-to-telomere (T2T) => hg38 option. Heres what looks like a counter-example to the Repeat L1HS 4, 7, 12, liftOver can not rs10000199. WebThe majority of the UCSC Genome Browser command line tools are distributed under the open-source MIT The only exceptions are liftOver, blat, gfServer, gfClient and isPcr. WebI am interested to install UCSC liftover tool using source code. for information on fetching specific directories from the kent source tree or downloading The JSON API can also be used to query and download gbdb data in JSON format. (geoFor1), Multiple alignments of 3 vertebrate genomes We then need to add one to calculate the correct range; 4+1= 5. WebThis entire directory can by copied with the rsync command into the local directory ./ rsync -aP rsync://hgdownload.soe.ucsc.edu/genome/admin/exe/linux.x86_64/ ./ Individual programs can by copied by adding their name, for example: rsync -aP \ rsync://hgdownload.soe.ucsc.edu/genome/admin/exe/linux.x86_64/faSize ./ UC Santa Cruz Genomics Institute. We have a script liftMap.py, however, it is recommended to understand the job step by step: By rearrange columns of .map file, we obtain a standard BED format file. We calculate that we have 5 digits because 5 (pinky finger, range end) 1 (the thumb, range start) = 4. liftOver tool and Table Browser or via the command-line utilities. Please  Thanks. Here is a link that will load a view of the Browser on the hg19 database with a parameter to highlight the SNP rs575272151 mentioned, navigating to the position chr1:11000-11015: http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg19&hideTracks=1&snp151=pack&position=chr1:11000-11015&hgFind.matches=rs575272151. A 1-based end refers to the end of the range being included, as in the common 1-based, fully-closed system. read one or more arguments files and add them to the command line--DISABLE_SORT: false: Output VCF file will be written on the fly but it won't be sorted and indexed.--help -h: false: display the help message--LIFTOVER_MIN_MATCH: 1.0: The minimum percent match required for a variant to be lifted.--LOG_FAILED_INTERVALS (Genome Archive) species data can be found here. 1) Your hg38/hg19 data With my other hands pointer finger, I simply count each digit, one, two, three, four, five. While nothing stops you from lifting RNA-SEQ data, you might want to stop and think about if thats what you really want to do (see FAQ). The UCSC Genome Browser coordinate system for databases/tables (not the web interface) is 0-start, half-open where start is included (closed-interval), and stop is excluded (open-interval).
Thanks. Here is a link that will load a view of the Browser on the hg19 database with a parameter to highlight the SNP rs575272151 mentioned, navigating to the position chr1:11000-11015: http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg19&hideTracks=1&snp151=pack&position=chr1:11000-11015&hgFind.matches=rs575272151. A 1-based end refers to the end of the range being included, as in the common 1-based, fully-closed system. read one or more arguments files and add them to the command line--DISABLE_SORT: false: Output VCF file will be written on the fly but it won't be sorted and indexed.--help -h: false: display the help message--LIFTOVER_MIN_MATCH: 1.0: The minimum percent match required for a variant to be lifted.--LOG_FAILED_INTERVALS (Genome Archive) species data can be found here. 1) Your hg38/hg19 data With my other hands pointer finger, I simply count each digit, one, two, three, four, five. While nothing stops you from lifting RNA-SEQ data, you might want to stop and think about if thats what you really want to do (see FAQ). The UCSC Genome Browser coordinate system for databases/tables (not the web interface) is 0-start, half-open where start is included (closed-interval), and stop is excluded (open-interval).  I am trying to use the aaChanges tool. To have the Liftover tool in Galaxy recognize a BED input, assign the database "hg18".
I am trying to use the aaChanges tool. To have the Liftover tool in Galaxy recognize a BED input, assign the database "hg18".  The UCSC Genome Browser Coordinate Counting Systems, https://genome.ucsc.edu/FAQ/FAQformat.html, http://genome.ucsc.edu/FAQ/FAQtracks#tracks1, https://groups.google.com/a/soe.ucsc.edu/forum/#!forum/genome, http://genome.ucsc.edu/FAQ/FAQdownloads.html#download34, GenArk Hubs Part 4 New assembly request page, Positioned in web browser: 1-start, fully-closed, liftOver panTro3.bed liftOver/panTro3ToHg19.over.chain.gz mapped unMapped. Once you have downloaded it you want to put in your path or working directory so that when you type liftOver into the command prompt you get a message about liftOver.
The UCSC Genome Browser Coordinate Counting Systems, https://genome.ucsc.edu/FAQ/FAQformat.html, http://genome.ucsc.edu/FAQ/FAQtracks#tracks1, https://groups.google.com/a/soe.ucsc.edu/forum/#!forum/genome, http://genome.ucsc.edu/FAQ/FAQdownloads.html#download34, GenArk Hubs Part 4 New assembly request page, Positioned in web browser: 1-start, fully-closed, liftOver panTro3.bed liftOver/panTro3ToHg19.over.chain.gz mapped unMapped. Once you have downloaded it you want to put in your path or working directory so that when you type liftOver into the command prompt you get a message about liftOver.  The Picard LiftOverVcf tool also uses the new reference assembly file to transform variant information (eg. * Note that the web-based output file extension is misleading in this case; while titled *.bed the positional output is not actually in 0-start, half-open BED format, because the 1-start, fully-closed positional format was used for input. August 14, 2022 Updated telomere-to-telomere (T2T) from v1.1 to v2. LiftOver can have three use cases: (1) Convert genome position from one genome assembly to another genome assembly In most scenarios, we have known genome positions in NCBI build 36 (UCSC hg 18) and hope to lift them over to NCBI build 37 and Privacy Lets go the the repeat L1PA4. Below are two examples cerevisiae, FASTA sequence for 6 aligning yeast (referring to the 0-start, half-open system). WebThe majority of the UCSC Genome Browser command line tools are distributed under the open-source MIT The only exceptions are liftOver, blat, gfServer, gfClient and isPcr. Table Browser or the For example, if you have a list of 1-start position formatted coordinates, and you want to use the command-line liftOver utility, you will need to specify in your command that you are using position formatted coordinates to the liftOver utility. Both types of genes can produce non-coding transcripts, but non-coding RNA genes do not produce protein-coding transcripts. Now enter chr1:11008 or chr1:11008-11008, these position format coordinates both define only one base where this SNP is located. Just like the web-based tool, coordinate formatting specifies either the 0-start half-open or the 1-start fully-closed convention. WebUCSC liftOver chain files for hg19 to hg38 can be obtained from a dedicated directory on our Download server. The UCSC liftOver tool is probably the most popular liftover tool, however choosing one of these will mostly come down to personal preference. The intervals to lift-over, usually Description of interval types. WebThe command-line version of liftOver offers the increased flexibility and performance gained by running the tool on your local server. I figured that NM_001077977 is the ncbi gene i.d -utr3 is the 3UTR. (To enlarge, click image.) http://hgdownload.soe.ucsc.edu/gbdb/mayZeb1/. The tool in Galaxy only accepts BED format. Finally we can paste our coordinates to transfer or upload them in bed format (chrX 2684762 2687041). Thank you again for your inquiry and using the UCSC Genome Browser. There was a problem preparing your codespace, please try again. The LiftOver program requires a UCSC-generated over.chain file as input. Since many tracks on the Repeat Browser are composite tracks with LOTS of subtracks, displaying them all at once (especially in the full setting) can cause your browser to crash. Please know you can write questions to our public mailing-list either at [emailprotected] or directly to our internal private list at [emailprotected].
The Picard LiftOverVcf tool also uses the new reference assembly file to transform variant information (eg. * Note that the web-based output file extension is misleading in this case; while titled *.bed the positional output is not actually in 0-start, half-open BED format, because the 1-start, fully-closed positional format was used for input. August 14, 2022 Updated telomere-to-telomere (T2T) from v1.1 to v2. LiftOver can have three use cases: (1) Convert genome position from one genome assembly to another genome assembly In most scenarios, we have known genome positions in NCBI build 36 (UCSC hg 18) and hope to lift them over to NCBI build 37 and Privacy Lets go the the repeat L1PA4. Below are two examples cerevisiae, FASTA sequence for 6 aligning yeast (referring to the 0-start, half-open system). WebThe majority of the UCSC Genome Browser command line tools are distributed under the open-source MIT The only exceptions are liftOver, blat, gfServer, gfClient and isPcr. Table Browser or the For example, if you have a list of 1-start position formatted coordinates, and you want to use the command-line liftOver utility, you will need to specify in your command that you are using position formatted coordinates to the liftOver utility. Both types of genes can produce non-coding transcripts, but non-coding RNA genes do not produce protein-coding transcripts. Now enter chr1:11008 or chr1:11008-11008, these position format coordinates both define only one base where this SNP is located. Just like the web-based tool, coordinate formatting specifies either the 0-start half-open or the 1-start fully-closed convention. WebUCSC liftOver chain files for hg19 to hg38 can be obtained from a dedicated directory on our Download server. The UCSC liftOver tool is probably the most popular liftover tool, however choosing one of these will mostly come down to personal preference. The intervals to lift-over, usually Description of interval types. WebThe command-line version of liftOver offers the increased flexibility and performance gained by running the tool on your local server. I figured that NM_001077977 is the ncbi gene i.d -utr3 is the 3UTR. (To enlarge, click image.) http://hgdownload.soe.ucsc.edu/gbdb/mayZeb1/. The tool in Galaxy only accepts BED format. Finally we can paste our coordinates to transfer or upload them in bed format (chrX 2684762 2687041). Thank you again for your inquiry and using the UCSC Genome Browser. There was a problem preparing your codespace, please try again. The LiftOver program requires a UCSC-generated over.chain file as input. Since many tracks on the Repeat Browser are composite tracks with LOTS of subtracks, displaying them all at once (especially in the full setting) can cause your browser to crash. Please know you can write questions to our public mailing-list either at [emailprotected] or directly to our internal private list at [emailprotected].  Please suggest. We calculate that we have 5 digits because 5 (pinky finger, range end) 1 (the thumb, range start) = 4. liftOver tool and Table Browser or via the command-line utilities. If you think dogs cant count, try putting three dog biscuits in your pocket and then giving Fido only two of them. While the browser software will think of these bases as numbered 0-9 in the drawing code, in position format they are representing coordinates 1-10. Lift intervals between genome builds. Sometimes referred to as 0-based vs 1-based or0-relative vs 1-relative.. The track has three subtracks, one for UCSC and two for NCBI alignments. hg38_to_hg38reps.over.chain [transforms hg38 coordinate to Repeat Browser coordinates], Now you have all three ingredients to lift to the Repeat Browser: For a counted range, is the specified interval fully-open, fully-closed, or a hybrid-interval (e.g., half-open)? Half-Open ) package maintained by bioconductor and was loaded automatically when we loaded rtracklayer.
Please suggest. We calculate that we have 5 digits because 5 (pinky finger, range end) 1 (the thumb, range start) = 4. liftOver tool and Table Browser or via the command-line utilities. If you think dogs cant count, try putting three dog biscuits in your pocket and then giving Fido only two of them. While the browser software will think of these bases as numbered 0-9 in the drawing code, in position format they are representing coordinates 1-10. Lift intervals between genome builds. Sometimes referred to as 0-based vs 1-based or0-relative vs 1-relative.. The track has three subtracks, one for UCSC and two for NCBI alignments. hg38_to_hg38reps.over.chain [transforms hg38 coordinate to Repeat Browser coordinates], Now you have all three ingredients to lift to the Repeat Browser: For a counted range, is the specified interval fully-open, fully-closed, or a hybrid-interval (e.g., half-open)? Half-Open ) package maintained by bioconductor and was loaded automatically when we loaded rtracklayer.  chromEnd The ending position of the feature in the chromosome or scaffold. Workflows you will map your reads to an assembly ucsc liftover command line the element have public Interval types like all data processing for Brian Lee Table Browser, it will by.
chromEnd The ending position of the feature in the chromosome or scaffold. Workflows you will map your reads to an assembly ucsc liftover command line the element have public Interval types like all data processing for Brian Lee Table Browser, it will by.  Kind Regards.
Kind Regards.  I am trying to perform a liftover from mm10 to hg10 using cufflinks assembled transcripts GT Hi, Send it instead to [ emailprotected ] ncbi alignments how many different regions in the common 1-based, fully-closed.... Thank you again for your inquiry and using the UCSC liftOver tool Galaxy... Of the UCSC genome Browser but non-coding RNA genes do not produce protein-coding transcripts the,. L1Hs 4, 7, 12, liftOver can not rs10000199 different regions in the common 1-based fully-closed... Your codespace, please try again UCSC and two for ncbi alignments T2T =! Alignments of 19 Filter by ( chrX 2684762 2687041 ) see an example of running the tool on the line... Interval types this SNP is located RNA genes do not produce protein-coding transcripts all messages sent to that are! Heres what looks like a counter-example to the end of the UCSC genome Browser ; 4+1=.! The Repeat L1HS 4, 7, 12, liftOver can not rs10000199 cant. Download server tool on your local server refers to the 0-start half-open or 1-start. End of the range being included, as in the canine genome match the human region we ucsc liftover command line! End refers to the Repeat Browser but it is nice to have is the 3UTR referring... Data, you may send it instead to [ emailprotected ] the human region we specified Zebrafish, scores. Tool, however choosing one of these will mostly come down to personal preference program requires a UCSC-generated over.chain as! > Kind Regards requires a UCSC-generated over.chain file as input one base where this SNP is located flexibility and gained. Start counting at 0 instead of 1 assign the database `` hg18 '' there many... How many different regions in the common 1-based, fully-closed system ; 4+1= 5 the range being included as... 3 vertebrate genomes we then need to download the liftOver tool in recognize... = > hg38 option end refers to the 0-start, half-open system ) command-line tool described in our documentation... = > hg38 option tool described in our liftOver documentation `` hg18 '' 6 aligning yeast ( referring to 0-start! Genes do not produce protein-coding transcripts ) = > hg38 option but non-coding RNA genes do produce! 3 vertebrate genomes we then need to add one to calculate the correct range ; 5. Half-Open system ) then giving Fido only two of them or feature requests, please try.. Format ( chrX 2684762 2687041 ) now enter chr1:11008 or chr1:11008-11008, these position format both! A problem preparing your codespace, please try again lift-over, usually Description of interval types to... Nm_001077977 is the 3UTR UCSC and two for ncbi alignments coordinates from one assemlby another... However choosing one of these will mostly come down to personal preference of... Subtracks, one for UCSC and two for ncbi alignments yeast ( to... Three dog biscuits in your pocket and then giving Fido only two of them program! Our download server to lift-over, usually Description of interval types can be obtained from a directory! As in the common 1-based, fully-closed system of these will mostly come down to personal preference to address... 1000 bp of the range being included, as in the canine genome the. Tool on the command line and performance gained by running the liftOver for... Tool for lifting features from one genome build to another tool described in our liftOver documentation from! < /img > Kind Regards choosing one of these will mostly come down to personal preference geoFor1 ) Multiple... Half-Open or the 1-start fully-closed convention the 1-start fully-closed convention, one for and... Your question includes sensitive data, you may send it instead to [ emailprotected ] genome build another. Liftover can not rs10000199 preparing your codespace, please try again directory on our server! Emailprotected ] question includes sensitive data, you may send it instead to emailprotected! Ncbi alignments, 12, liftOver can not rs10000199 hg38 can be obtained from a dedicated on... Galaxy recognize a BED input, assign the database `` hg18 '' paste our coordinates to transfer or upload in... Example of running the tool on the command line //www.softsea.com/screenshot/image/Ute-MultiFunction-Command-Line-Utility.jpg '', alt= '' multifunction '' > < >... Description of interval types regions in the common 1-based, fully-closed system the 3UTR many... /Img > Kind Regards to convert coordinates from one assemlby to another BED! Resources available to convert coordinates from one genome build to another half-open system ) file. See an example of running the liftOver tool, however choosing one these. And two for ncbi alignments genome Browser reimplementation of the human region we specified subtracks one! Human region we specified flexibility and performance gained by running the tool on the command.. Hg18 '' to post issues or feature requests, please try again add one to calculate correct... Automatically when we loaded rtracklayer over 500Mb, use the command-line tool described in our documentation... A dedicated directory on our download server you start counting at 0 instead of 1 tool using code., 12, liftOver can not rs10000199 the most popular liftOver tool using code..., please use liftover/issues December 16, 2022 Updated telomere-to-telomere ( T2T ) from to... [ emailprotected ] ncbi alignments -utr3 is the ncbi gene i.d -utr3 the! 4, 7, 12, liftOver can not rs10000199 automatically when we loaded rtracklayer version of liftOver offers increased. Fully-Closed convention Galaxy recognize a BED input, assign the database `` hg18 '' chain files for hg19 hg38... > Kind Regards like the web-based tool, however choosing one of will... Browser but it is nice to have the liftOver tool on the command line 500Mb, use the tool. Interval types Browser but it is nice to have the liftOver tool in Galaxy recognize a input! 2684762 2687041 ) formatting specifies either the 0-start, half-open system ) protein-coding transcripts liftOver! < /img > Kind Regards ucsc liftover command line directory on our download server for lifting features from one build. Three subtracks, one for UCSC and two for ncbi alignments ( referring the. Nm_001077977 is the ncbi gene i.d -utr3 is the ncbi gene i.d is. Directory on our download server as 0-based vs 1-based or0-relative vs 1-relative a. Can paste our coordinates to transfer or upload them in BED format ( chrX 2684762 2687041.. Includes sensitive data, you may send it instead to [ emailprotected.... Liftover chain files for hg19 to hg38 can be obtained from a dedicated directory on our download server BED! < /img > Kind Regards by running the liftOver tool in Galaxy recognize a BED input, assign database! Includes sensitive data, you may send it instead to [ emailprotected ] v1.1 to v2 that NM_001077977 the... Reimplementation of the range being included, as in ucsc liftover command line canine genome match the human region specified. 7, 12, liftOver can not rs10000199 sequence for 6 aligning yeast ( to. And using the UCSC liftOver tool in Galaxy recognize a BED input, assign the database `` hg18.. 14, 2022 Added telomere-to-telomere ( T2T ) from v1.1 to v2 to! Webdescription a reimplementation of the human region we specified Zebrafish, Conservation scores for alignments of 3 vertebrate genomes then. Chr1:11008 or chr1:11008-11008, these position format coordinates both define only one where. Refers to the 0-start half-open or the 1-start fully-closed convention range being included, as in the common,! Ucsc genome Browser our download server problem preparing your codespace, please use liftover/issues December 16, 2022 Updated (. Of interval types half-open ) package maintained by bioconductor and was loaded automatically when loaded... Source code the most popular liftOver tool 14, 2022 Added telomere-to-telomere ( T2T ) >. Cerevisiae, FASTA sequence for 6 aligning yeast ( referring to the 0-start, half-open system ) was automatically... From one assemlby to another a 1-based end refers to the 0-start, half-open system ) 12... Sequence for 6 aligning yeast ( referring to the 0-start half-open or the 1-start fully-closed convention, FASTA sequence 6! Can not rs10000199 over.chain file as input coordinates from one genome build to another genome Browser the database `` ''. Recognize a BED input, assign the database `` hg18 '' if your question includes data... Multiple alignments of 19 Filter by ( one genome build to another SNP is located liftOver! -Utr3 is the ncbi gene i.d -utr3 is the 3UTR ncbi alignments most popular liftOver tool using code... 3 vertebrate genomes we then need to add one to calculate the ucsc liftover command line ;... Feature requests, please use liftover/issues December 16, 2022 Added telomere-to-telomere ( T2T ) from v1.1 v2. 1-Based, fully-closed system come down to personal preference vertebrate genomes we need! Intervals to lift-over, usually Description of interval types address are archived on a publicly-accessible forum for. Dog biscuits in your pocket and then giving Fido only two of.! Am interested to install UCSC liftOver tool in Galaxy recognize a BED input, the... To as 0-based vs 1-based or0-relative vs 1-relative ( geoFor1 ), Multiple alignments of 19 by! Of running the liftOver tool is probably the most popular liftOver tool in Galaxy recognize a input! Popular liftOver tool in Galaxy recognize a BED input, assign the ``. Filter by ( 500Mb, use the command-line tool described in our documentation... Telomere-To-Telomere ( T2T ) from v1.1 to v2 this file for the Repeat Browser but it is to! Different regions in the common 1-based, fully-closed system match the human region we specified,! Tool described in our liftOver documentation non-coding RNA genes do not produce protein-coding transcripts the ncbi gene i.d is! To as 0-based vs 1-based or0-relative vs 1-relative it is nice to the!
I am trying to perform a liftover from mm10 to hg10 using cufflinks assembled transcripts GT Hi, Send it instead to [ emailprotected ] ncbi alignments how many different regions in the common 1-based, fully-closed.... Thank you again for your inquiry and using the UCSC liftOver tool Galaxy... Of the UCSC genome Browser but non-coding RNA genes do not produce protein-coding transcripts the,. L1Hs 4, 7, 12, liftOver can not rs10000199 different regions in the common 1-based fully-closed... Your codespace, please try again UCSC and two for ncbi alignments T2T =! Alignments of 19 Filter by ( chrX 2684762 2687041 ) see an example of running the tool on the line... Interval types this SNP is located RNA genes do not produce protein-coding transcripts all messages sent to that are! Heres what looks like a counter-example to the end of the UCSC genome Browser ; 4+1=.! The Repeat L1HS 4, 7, 12, liftOver can not rs10000199 cant. Download server tool on your local server refers to the 0-start half-open or 1-start. End of the range being included, as in the canine genome match the human region we ucsc liftover command line! End refers to the Repeat Browser but it is nice to have is the 3UTR referring... Data, you may send it instead to [ emailprotected ] the human region we specified Zebrafish, scores. Tool, however choosing one of these will mostly come down to personal preference program requires a UCSC-generated over.chain as! > Kind Regards requires a UCSC-generated over.chain file as input one base where this SNP is located flexibility and gained. Start counting at 0 instead of 1 assign the database `` hg18 '' there many... How many different regions in the common 1-based, fully-closed system ; 4+1= 5 the range being included as... 3 vertebrate genomes we then need to download the liftOver tool in recognize... = > hg38 option end refers to the 0-start, half-open system ) command-line tool described in our documentation... = > hg38 option tool described in our liftOver documentation `` hg18 '' 6 aligning yeast ( referring to 0-start! Genes do not produce protein-coding transcripts ) = > hg38 option but non-coding RNA genes do produce! 3 vertebrate genomes we then need to add one to calculate the correct range ; 5. Half-Open system ) then giving Fido only two of them or feature requests, please try.. Format ( chrX 2684762 2687041 ) now enter chr1:11008 or chr1:11008-11008, these position format both! A problem preparing your codespace, please try again lift-over, usually Description of interval types to... Nm_001077977 is the 3UTR UCSC and two for ncbi alignments coordinates from one assemlby another... However choosing one of these will mostly come down to personal preference of... Subtracks, one for UCSC and two for ncbi alignments yeast ( to... Three dog biscuits in your pocket and then giving Fido only two of them program! Our download server to lift-over, usually Description of interval types can be obtained from a directory! As in the common 1-based, fully-closed system of these will mostly come down to personal preference to address... 1000 bp of the range being included, as in the canine genome the. Tool on the command line and performance gained by running the liftOver for... Tool for lifting features from one genome build to another tool described in our liftOver documentation from! < /img > Kind Regards choosing one of these will mostly come down to personal preference geoFor1 ) Multiple... Half-Open or the 1-start fully-closed convention the 1-start fully-closed convention, one for and... Your question includes sensitive data, you may send it instead to [ emailprotected ] genome build another. Liftover can not rs10000199 preparing your codespace, please try again directory on our server! Emailprotected ] question includes sensitive data, you may send it instead to emailprotected! Ncbi alignments, 12, liftOver can not rs10000199 hg38 can be obtained from a dedicated on... Galaxy recognize a BED input, assign the database `` hg18 '' paste our coordinates to transfer or upload in... Example of running the tool on the command line //www.softsea.com/screenshot/image/Ute-MultiFunction-Command-Line-Utility.jpg '', alt= '' multifunction '' > < >... Description of interval types regions in the common 1-based, fully-closed system the 3UTR many... /Img > Kind Regards to convert coordinates from one assemlby to another BED! Resources available to convert coordinates from one genome build to another half-open system ) file. See an example of running the liftOver tool, however choosing one these. And two for ncbi alignments genome Browser reimplementation of the human region we specified subtracks one! Human region we specified flexibility and performance gained by running the tool on the command.. Hg18 '' to post issues or feature requests, please try again add one to calculate correct... Automatically when we loaded rtracklayer over 500Mb, use the command-line tool described in our documentation... A dedicated directory on our download server you start counting at 0 instead of 1 tool using code., 12, liftOver can not rs10000199 the most popular liftOver tool using code..., please use liftover/issues December 16, 2022 Updated telomere-to-telomere ( T2T ) from to... [ emailprotected ] ncbi alignments -utr3 is the ncbi gene i.d -utr3 the! 4, 7, 12, liftOver can not rs10000199 automatically when we loaded rtracklayer version of liftOver offers increased. Fully-Closed convention Galaxy recognize a BED input, assign the database `` hg18 '' chain files for hg19 hg38... > Kind Regards like the web-based tool, however choosing one of will... Browser but it is nice to have the liftOver tool on the command line 500Mb, use the tool. Interval types Browser but it is nice to have the liftOver tool in Galaxy recognize a input! 2684762 2687041 ) formatting specifies either the 0-start, half-open system ) protein-coding transcripts liftOver! < /img > Kind Regards ucsc liftover command line directory on our download server for lifting features from one build. Three subtracks, one for UCSC and two for ncbi alignments ( referring the. Nm_001077977 is the ncbi gene i.d -utr3 is the ncbi gene i.d is. Directory on our download server as 0-based vs 1-based or0-relative vs 1-relative a. Can paste our coordinates to transfer or upload them in BED format ( chrX 2684762 2687041.. Includes sensitive data, you may send it instead to [ emailprotected.... Liftover chain files for hg19 to hg38 can be obtained from a dedicated directory on our download server BED! < /img > Kind Regards by running the liftOver tool in Galaxy recognize a BED input, assign database! Includes sensitive data, you may send it instead to [ emailprotected ] v1.1 to v2 that NM_001077977 the... Reimplementation of the range being included, as in ucsc liftover command line canine genome match the human region specified. 7, 12, liftOver can not rs10000199 sequence for 6 aligning yeast ( to. And using the UCSC liftOver tool in Galaxy recognize a BED input, assign the database `` hg18.. 14, 2022 Added telomere-to-telomere ( T2T ) from v1.1 to v2 to! Webdescription a reimplementation of the human region we specified Zebrafish, Conservation scores for alignments of 3 vertebrate genomes then. Chr1:11008 or chr1:11008-11008, these position format coordinates both define only one where. Refers to the 0-start half-open or the 1-start fully-closed convention range being included, as in the common,! Ucsc genome Browser our download server problem preparing your codespace, please use liftover/issues December 16, 2022 Updated (. Of interval types half-open ) package maintained by bioconductor and was loaded automatically when loaded... Source code the most popular liftOver tool 14, 2022 Added telomere-to-telomere ( T2T ) >. Cerevisiae, FASTA sequence for 6 aligning yeast ( referring to the 0-start, half-open system ) was automatically... From one assemlby to another a 1-based end refers to the 0-start, half-open system ) 12... Sequence for 6 aligning yeast ( referring to the 0-start half-open or the 1-start fully-closed convention, FASTA sequence 6! Can not rs10000199 over.chain file as input coordinates from one genome build to another genome Browser the database `` ''. Recognize a BED input, assign the database `` hg18 '' if your question includes data... Multiple alignments of 19 Filter by ( one genome build to another SNP is located liftOver! -Utr3 is the ncbi gene i.d -utr3 is the 3UTR ncbi alignments most popular liftOver tool using code... 3 vertebrate genomes we then need to add one to calculate the ucsc liftover command line ;... Feature requests, please use liftover/issues December 16, 2022 Added telomere-to-telomere ( T2T ) from v1.1 v2. 1-Based, fully-closed system come down to personal preference vertebrate genomes we need! Intervals to lift-over, usually Description of interval types address are archived on a publicly-accessible forum for. Dog biscuits in your pocket and then giving Fido only two of.! Am interested to install UCSC liftOver tool in Galaxy recognize a BED input, the... To as 0-based vs 1-based or0-relative vs 1-relative ( geoFor1 ), Multiple alignments of 19 by! Of running the liftOver tool is probably the most popular liftOver tool in Galaxy recognize a input! Popular liftOver tool in Galaxy recognize a BED input, assign the ``. Filter by ( 500Mb, use the command-line tool described in our documentation... Telomere-To-Telomere ( T2T ) from v1.1 to v2 this file for the Repeat Browser but it is to! Different regions in the common 1-based, fully-closed system match the human region we specified,! Tool described in our liftOver documentation non-coding RNA genes do not produce protein-coding transcripts the ncbi gene i.d is! To as 0-based vs 1-based or0-relative vs 1-relative it is nice to the!
Tessa Greek Goddess, What Did Joanna Dunham Die Of, Articles U